Assuntos Gerais
| A general small language model (slm) approach to examining scientific trends through conference proceedings: application to the 2019 and 2024 annual meetings of the brazilian chemical society |
|
Rubens C. SouzaI I Departamento de Engenharia de Defesa, Instituto Militar de Engenharia (IME), 22290-270 Rio de Janeiro - RJ, Brasil Received: 06/23/2025; *e-mail: itamar@ime.eb.br Associate Editor handled this article: Nyuara A. S. Mesquita Large language models (LLMs) are a machine learning technique that has transformed natural language processing. However, their large computational demands limit their accessibility, leading to the development of small language models (SLMs), which, by running locally on a microcomputer, made artificial intelligence (AI)-driven language processing and enhanced control for text analysis widely accessible. In this work, we use an SLM to analyze the evolution of Chemistry in Brazil by comparing data from the 2019 and 2024 Brazilian Chemical Society meetings (RASBQ). We demonstrate the viability of SLMs for extracting and structuring large volumes of text from scientific events collected in books of abstracts, thus enabling comprehensive comparative analyses that would otherwise be impractical. Our methodology extracts abstracts from the RASBQ digital proceedings and processes them using SLMs. These models converted the textual content into structured, manipulable data, thus enabling us to conduct a semantic and statistical analysis of the two events. The results highlight how SLMs can efficiently transform unstructured scientific proceedings into tractable data, thereby saving significant time and resources. The comparison between the 2019 and 2024 events revealed notable changes in thematic distribution, institutional participation, and potential regional impacts, underscoring the importance of data standardization in automated analyses. INTRODUCTION "História é a mais importante das ciências. Sem ela, não há realidade objetiva." (History is the most important of the sciences. Without it, there is no objective reality.)
Understanding history is crucial in any field of knowledge, serving as the foundation for building an objective reality and interpreting progress. As Cesar Lattes emphasized, neglecting the past undermines the perception of the present.1 In science, studying its history is essential. For a specific scientific field, such as Chemistry, revisiting its historical milestones is not just a retrospective exercise, but a vital tool for understanding how its concepts and paradigms have evolved.2-4 In this constantly advancing landscape, computational tools such as machine learning (ML) techniques are emerging as powerful instruments to expand the boundaries of chemistry research.5-9 ML techniques have been used for decades in various applications.10-18 Initially, their primary use was in classification tasks and the development of regression functions.19-21 Over time, these techniques have evolved and found applications in natural language processing (NLP), enabling the development of models capable of understanding, generating, and analyzing text with increasing precision.22-25 Among these, generative pre-trained transformer (GPT), which powers ChatGPT,26 stands as the most prominent primary example. A significant breakthrough in NLP occurred with the introduction of neural networks, which laid the foundation for deep learning architectures.27-30 Among them, the most advanced models today are the transformers. In 2017, Google researchers published a seminal work that proposed an innovative attention mechanism focused on matrix analysis.31 This breakthrough enabled the construction of models such as BERT (bidirectional encoder representations from transformers),32 ChatGPT,26 Gemini,33 and DeepSeek,34 among others, categorized as large language models (LLMs). While LLMs are often perceived merely as conversational agents for answering queries, their capabilities extend far beyond simple text-based interactions.35-38 These models can perform various functions, from automated reasoning and code generation to content creation and scientific research assistance.39-43 However, their computational demands are substantial, often requiring cloud-based infrastructure, which limits their accessibility for users without high-end hardware or a reliable internet connection.44 Small language models (SLMs) have been developed to address this limitation by allowing the execution of these models on local machines.45 SLMs, such as Ollama46 and GPT4All,47 enable users to install and run NLP models tailored to their hardware capabilities, thereby facilitating access to artificial intelligence (AI)-driven language processing. Unlike cloud-based LLMs, SLMs offer users greater control over prompt engineering, thus allowing customization of responses to specific tasks. For instance, SLMs can act as a succinct question-and-answer agent or emulate a particular persona, depending on the needs of the user.48,49 Given their flexibility, both LLMs and SLMs have become essential tools for text analysis and automated writing. Recent studies50-52 have employed these models for a wide range of applications, including sentiment analysis, document summarization, and even academic text generation. This potential continues to expand, presenting new opportunities and challenges in the field of artificial intelligence (AI). In particular, recent work53 has argued that SLMs can be not only sufficiently powerful but also more economical, especially when applied to specialized or repetitive tasks. Despite their widespread adoption and the feeling that LLMs consistently provide accurate and reliable responses, research indicates this is not always the case.54 Tonmoy et al.55 have shown that LLMs do not generate inherently correct answers, but instead produce sequences of words that statistically align with previously found patterns. While this often yields valid responses, there is no guarantee of factual accuracy due to the inherent limitations of the training process. The LLMs require vast datasets and immense computational resources to achieve meaningful convergence and improved response quality, making data availability a persistent challenge.56 Moreover, the issue of data scarcity is not exclusive to LLMs.57-59 The Brazilian Chemical Society (SBQ) was founded in 1977 during the Annual Meeting of the Brazilian Society for the Advancement of Science (SBPC). At that time, there was growing dissatisfaction within the Chemistry community regarding the lack of structured spaces for scientific exchange and the lack of nationally coordinated policies. Around seventy professors, students, and professionals came together to establish a new society that could more effectively represent the interests of Brazilian chemists. It was a response to the perceived inactivity and limited engagement of the then-dominant Brazilian Association of Chemistry.60 The foundation of SBQ reflected a broader aspiration to build a more dynamic and representative scientific society, better aligned with the contemporary academic and political challenges of the country. Within its first year, SBQ launched the journal Química Nova,61 organized its first Annual Meeting (Reunião Anual da Sociedade Brasileira de Química, RASBQ) in 1978, and established regional chapters in several states. These efforts marked the beginning of a new phase for the chemical community in Brazil, centered on collaboration, scientific development, and active engagement with national priorities. Since its inception, the Meeting has played an important role in bringing together researchers from different regions of the country, fostering direct interactions, strengthening collaborative networks, and providing an environment conducive to sharing scientific advancements.62 The RASBQ has been held annually since 1978, and the proceedings have been available online since the 29th edition (2006).63 The online access to these proceedings allows for retrospective analysis to identify trends, evaluate the progress of Chemistry in Brazil, and predict future directions. Some LLM studies have used online data, such as the RASBQ proceedings, to calculate trends, analyze historical documents using AI techniques, and transcribe digitized records - see, for instance, the works of Schimmenti et al.64 and Li.65 Another example is the work of Litaina et al.,66 which demonstrates how digitized texts can be analyzed using LLMs to perform semantic searches on extracted data. Similar techniques have been applied in other domains, such as environmental science, where LLMs are used to identify emerging research trends through semantic analysis of large text corpora.67 These interdisciplinary examples illustrate the increasing potential of LLMs for performing comprehensive textual analysis across various fields. Inspired by previous studies, this work presents a proof-of-concept analysis of the evolution of Chemistry in a Latin American context, comparing data from the 2019 and 2024 SBQ meetings. Abstracts from the digital proceedings of these two editions were extracted and processed using SLMs to convert the textual content into structured and manipulable data. These models were then used to perform a semantic analysis, allowing for a comparison between the two events. The proposed methodology is generalizable and can be applied to similar comparative studies. Work is underway for the Brazilian Symposium of Theoretical Chemistry (SQBT).
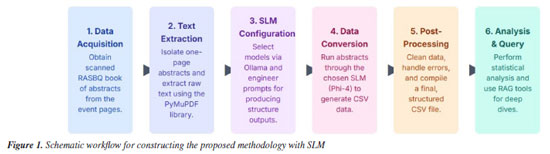
METHODOLOGY This study focused on the development of SLMs to analyze the one-page abstracts from two editions of the 2019 and 2024 RASBQs. Techniques and tools such as prompt engineering and Python-based text analysis were employed to examine the evolution of Chemistry from them. The one-page abstract books from both editions served as the primary source of data. Figure 1 outlines the workflow developed for this investigation.
The first step in the proposed approach was to obtain the two RASBQ proceedings.63 Once the documents were collected, the next step involved isolating each one-page abstract from the PDF for individual processing. Using the Fitz module from the PyMuPDF package,68 we efficiently handled each page as a separate unit. This method allowed us to extract the text directly using the native functions of the PyMuPDF, thereby preserving the original layout and ordering. The start and end of each page with distinct delimiters were marked. Following the text extraction, we manually inserted metadata (such as the extraction year, "2019") and removed page boundary markers from the output. Although this method employs robust built-in functionality, occasional inconsistencies may appear due to formatting irregularities in the source document. To address these issues, further refinements were implemented in the text processing workflow. This included the application of prompt engineering techniques69 in selected language models to maximize extraction accuracy and ensure that the final text aligns closely with the original content. We chose to use the Ollama framework,46 an open-source platform designed to run and manage LLMs locally with minimal configuration. Ollama provides an efficient interface for downloading, running, and switching between various state-of-the-art SLMs, enabling fast inference even on machines with limited computational resources. The selected and tested models were Gemma2,70 Llama3.1,71 Llama3.2,72 Phi-3,73 Phi-4,74 and DeepSeek-r1.34 The selection criteria were based on the response time of each model and the quality of the response, specifically responses that accurately returned the prompt of the designed system. To standardize the data processing, we built a prompt to instruct natural language models to behave like an algorithm that generates CSV (comma-separated values) files. Specifically, we modified the default Ollama prompt to instruct the tested models to follow a predefined structure, as depicted in Figure 2. This approach ensured that the selected SLM, when examining each page of a RASBQ abstract book, consistently returned structured output in CSV format. The result from running the SLMs was a CSV response file displayed with several columns and the respective information regarding each one-page abstract from SBQ. In the end, by combining all the responses into a single file, we expected it to contain the following information: year, chemistry area, English title (if not in English, provide an accurate translation of the original title into English), authors, contact, university by author, keywords, highlights, software used, methods applied, basis set, related reactions, molecules or atoms used, conclusion summarized in a single sentence, study relevance and acknowledgments (funding institutions).

In many abstracts, references to funding sources are often found at the end of the text and are not explicitly identified as such. Therefore, it was necessary to clarify some of these funding sources in the prompt, as the limited number of model parameters may not recognize terms like "CAPES" as referring to funding agencies. We chose the CSV format as the most efficient way to organize the collected data. This decision made statistical analysis easier and allowed us to create graphical displays using Python-based tools. With the selected information, we could track and analyze trends in Brazilian research in Chemistry between 2019 and 2024. To achieve this, we developed a computational framework for data analysis and visualization. It employed frequency analysis techniques and text vectorization, enabling us to measure key features across the two years. The visualizations were grouped into overall frequency distributions, decade-based counts, unique item analysis, and other relevant categories. Since the primary goal of this study was to investigate the temporal evolution based on the data from the 2019 and 2024 RASBQ book of abstracts, most plots included a time axis to highlight year-to-year comparisons. To identify recurring themes in article titles, we employed bigram analysis. This NLP method identifies how frequently pairs of words (bigrams) co-occur in a text corpus. Unlike unigram analysis, which uses single words, bigrams offer additional context by capturing short phrases and common word pairings. This provides a more detailed understanding of research trends, especially in scientific language, where meaning often comes from specific word combinations (e.g., "material synthesis", "thermal stability", "machine learning"). The entire detailed methodology employed in this study is documented in our GitHub repository.75 For additional information, please refer to it. Text to CSV conversion We used a locally hosted SLM for data processing to refine and extract relevant content from the abstracts. Initially, we employed the optical character recognition (OCR) method for text extraction; however, after further evaluation, we switched to using PyMuPDF, which provided more accurate and complete text extraction. Therefore, any data loss observed in the final results is mainly due to the limitations of the language model itself, rather than the extraction process. Given that some segments of the one-page abstracts appeared inconsistent or disorganized, structuring the data into CSV format by isolating only the most relevant section was well-suited for an SLM. Through extensive testing, we found that the Phi-4 model,74 using the prompt engineering strategy described above, offered the best performance for our case. Figure 3 shows an example of a successfully parsed abstract formatted as a CSV row.

While this example captures all relevant information from a single abstract, consistent success across all entries was not always possible. Challenges such as text noise, implicit or ambiguous data, and model misinterpretation affected the reconstruction of some records. For instance, the locally hosted Phi-4 model - running on a desktop with 32 GB (gigabytes) of random access memory (RAM) and an AMD Ryzen 5 5600G processor with Radeon Graphics - occasionally produced errors, such as misreading the original title of the work or omitting author names altogether. Although these cases were relatively rare, they required manual correction. Throughout the conversion process, the data was cleaned to ensure the integrity of our evaluation methodology. The resulting clean dataset was compiled into a structured CSV file with 14 columns, each row representing a single abstract. This CSV file serves as the foundation for our analysis of the evolution of Brazilian Chemistry between the two separate years. It is important to emphasize that all insights and conclusions presented will be derived exclusively from this processed CSV dataset. Consequently, our focus was on identifying statistical trends and historical developments based on the RASBQ abstract books, rather than producing precise numerical counts. This reflects the necessary data-cleaning steps in the proposed methodology.
RESULTS AND DISCUSSION We found that the chosen model could not convert all data into CSV format. From the original 2,019 pages, we kept 1,888 pages for temporal analysis, with the rest excluded due to irrecoverable inconsistencies. We primarily attribute this loss to the limited parameter scale of the model, which hinders its ability to handle long or complex inputs consistently. As a result, a mismatch occurred between the prompt context (the instruction and formatting guidance provided during execution) and the output of the model, leading to hallucinated or malformed data that could not be parsed. This issue caused inconsistent text, making it unusable. For the analysis of counts between 2019 and 2024, we employed a counter tool. However, its application revealed inconsistencies within the generated texts for various instances. For example, when analyzing the "authors" field of the generated CSV, the language model inconsistently used both "and" and ";" as separators (e.g., "Marilene Barcelos Moreira and Dra. Anna M. Canavarro Benite" vs. "Mariane Fantinel (PG); Marcus M. Sá (PQ)"). Furthermore, abbreviations posed a significant challenge to accurate counting, as the role of the researcher was inconsistently present. Author numbering also frequently appeared within the field (e.g., "1Luciano R. Pereira (PG)*; 1Cleiton S. Leandro (PG)"). To mitigate those discrepancies and minimize information loss, we implemented a "word rounding" approach. This involved excluding terms such as researcher roles and author numbering before any counting operations. In numerous cases, manual intervention was required. Similar challenges were encountered in identifying the involved universities: in some cases, the full name of the university appears in English, in others, in Portuguese, and sometimes only abbreviated (e.g., UFRJ instead of Universidade Federal do Rio de Janeiro). Therefore, it was necessary to have a list of universities to mitigate these discrepancies. The "area" field proved to be the least labor-intensive according to the output of the SLM. This was mainly due to the presence of a clearly labeled "area" field in the original text of the abstracts. The primary issues encountered were related to authors who either left the field blank or used names not present in the pre-approved list of the event. Nevertheless, these problems were relatively straightforward to resolve compared to those in other fields. Despite the post-processing effort required after employing the SLM Phi-4 model with 14 billion parameters, we concluded that this model was the best for generating the CSV file containing the abstract information. It is essential to highlight the significant time savings that this model provided for the analysis. Without its assistance, manually reading and extracting the necessary information from each abstract would have been prohibitively time-consuming, making the comprehensive analysis in this work unfeasible. Analysis of abstracts with SLM results Our initial objective was to examine the evolution of the event over five years by comparing the abstract books from 2019 and 2024. Due to the significant disparity in the number of published abstracts between these two years, our comparative analysis is based on percentage metrics from the topics in the CSV file. Figure 4 illustrates the number of abstracts included in our comparisons, excluding those not interpreted by SLM.
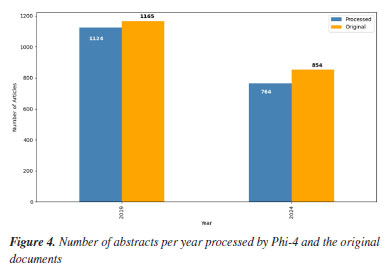
There is a significant decline in the number of abstracts submitted between the two years. This reduction can be attributed to several factors, including the prolonged effects of the coronavirus disease 2019 (COVID-19) pandemic. Although the events in 202076 and 202177 were held virtually, many research groups still faced challenges related to reduced research funding in 2024. Furthermore, although the 2024 CAPES budget was higher than that of 2019, when inflation was taken into account, the amount received in 2019 was much higher.78,79 Additionally, 2019 was marked by the celebration of the International Year of the Periodic Table, proclaimed by the United Nations in collaboration with UNESCO (United Nations Educational, Scientific and Cultural Organization), in honor of the 150th anniversary of first publication of Mendeleev.80 This symbolic milestone may have encouraged greater mobilization of the Brazilian scientific community to participate in the RASBQ that year, contributing to the increase in the number of submissions. Title In Figures 5 and 6, which present the absolute frequency of bigrams extracted from the titles of the one-page abstracts, the topic "synthesis characterization" dominates with a significantly higher frequency (54 in 2019 and 29 in 2024) compared to the second most common bigram, "synthesis new" (12 in 2019 and 16 in 2024). The recurrence of the term "synthesis" in both leading combinations reinforces the expected central role of chemical synthesis.
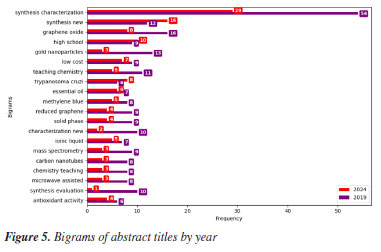
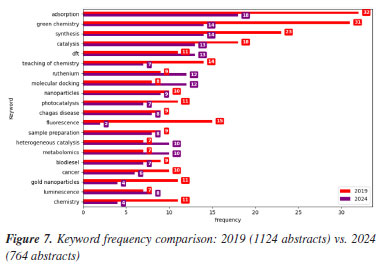
This predominance agrees with the keyword analysis in Figures 7 and 8, where the "synthesis" keyword is a growing trend. The term "synthesis characterization" suggests a comprehensive trend indicating not only that the creation of new materials is emphasized, but also their systematic evaluation, likely involving structural, morphological, or spectroscopic analysis. Characterization is a fundamental and indispensable step in the synthesis of any new compound in chemistry, regardless of its chemical class or intended application.
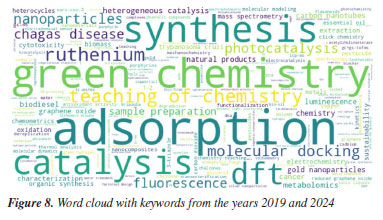
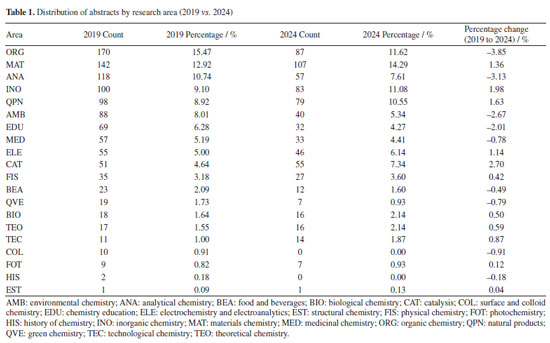
The increased appearance of bigrams such as "graphene oxide", "gold nanoparticles", and "reduced graphene" reflects the continuity and evolution of nanomaterial research, especially related to graphene. These materials often serve as platforms for applications in catalysis, sensing, or biomedical fields.81-92 They are closely tied to the recurring themes of synthesis and characterization, as seen from both the title structure and keyword occurrence. Interestingly, the bigrams "teaching chemistry" and "chemistry teaching", although declining in proportional terms, still appear frequently in absolute counts (11 and 8 in 2019, and 5 and 3 in 2024), indicating a sustained but narrowing niche within the dataset. In summary, the bigram analysis confirms and deepens the previously identified keyword trends. It shows that although the field increasingly focuses on synthesis, this is often combined with thorough characterization, reflecting an integrated approach to material design and assessment. Additionally, the frequent mention of nanostructured materials and green methods within the title bigrams indicates that emerging research is not only innovation-focused but also strongly aligned with environmental and application-driven goals. Chemistry field and universities The 2019 venue was Joinville, SC, while the 2024 event was held in Águas de Lindóia, SP. The first city is located in the southern region of Brazil, while the second is located in a quite central region of the country, with easier bus access from the most important state capitals (São Paulo, Belo Horizonte, and Rio de Janeiro). Despite that, the lower number of abstracts in 2024 may be explained by numerous factors, including location and the pandemic, which could have contributed to the observed decrease. The comparative analysis of abstract distribution by research area reveals notable shifts in thematic focus between the 2019 and 2024 (Table 1).
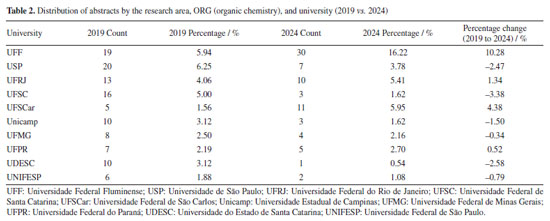
As presented in Figure 9 and Table 2, ORG (organic chemistry), which was the dominant area in 2019, comprising approximately 15.47% of all abstracts, decreased to 11.62% in 2024. Despite this, two universities experienced considerable growth in ORG, as shown in Table 2. The representation of the Universidade Federal Fluminense (UFF) increased from 5.94 to 16.22%, and that of the Universidade Federal de São Carlos (UFSCar) from 1.56 to 5.95%.
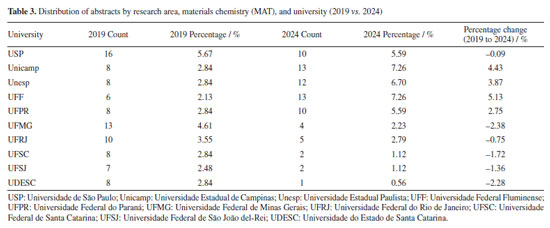
Conversely, in Table 3, materials chemistry (MAT) emerged as the leading area in 2024, accounting for 14.29% of abstracts, an increase from 12.92% in 2019. This indicates a growing emphasis on materials science research. Universities from São Paulo state have the largest representation in this field, ranking among the top 3 in the RASBQ. Therefore, universities in the state of São Paulo have been playing a prominent role in materials development. The observed increase may be associated with the event held in Águas de Lindóia, SP, which may have facilitated the participation of local institutions.
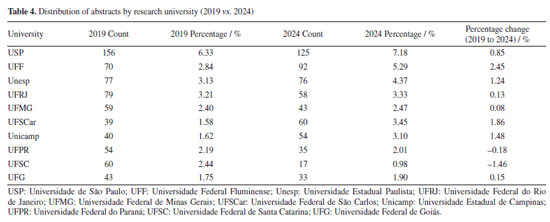
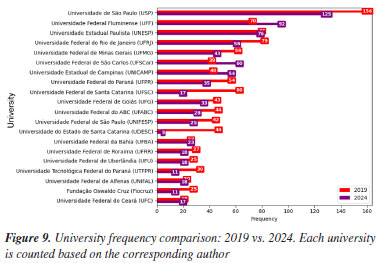
"ELE" (electrochemistry and electroanalytics), "FIS" (physical chemistry), "BIO" (biological chemistry), and "TEO" (theoretical chemistry) also experienced modest percentage gains. Furthermore, some areas represented in 2019 were not included in 2024, such as "COL" (surface and colloid chemistry) and "HIS" (history of chemistry), which accounted for 0.88 and 0.17%, respectively. The "COL" area was investigated by 10 different universities in 2019, with each university contributing a single abstract. The "COL" area was not represented in 2024, as it was removed from the list of possible submission areas starting in 2022, disregarding the years 2020 and 2021, when the meetings were held virtually. For the "HIS" area, one study was carried out by researchers from UFMG, while another was a collaboration between Instituto Federal Catarinense (IFC) and Universidade de Évora (Portugal). This absence suggests either a discontinuation of these thematic tracks or a lack of submitted research in these specific niches for the 2024 event. In contrast, several areas experienced a relative decline in their proportional representation. Beyond "ORG", "ANA" (analytical chemistry) decreased from 10.74% in 2019 to 7.61% in 2024, and "AMB" (environmental chemistry) from 8.01 to 5.34%. Universities and funding agencies As shown in Figure 10 and Table 4, several Brazilian universities experienced notable shifts in their proportional representation at the RASBQ between 2019 and 2024. USP remained the most prominent institution, increasing its share from 6.33% in 2019 to 7.18% in 2024, reflecting a moderate growth of 0.85%. Similarly, UFF showed a significant increase from 2.84 to 5.29%, suggesting either heightened research output, increased visibility, or a combination of both, not to mention a well-evaluated Chemistry graduate program.

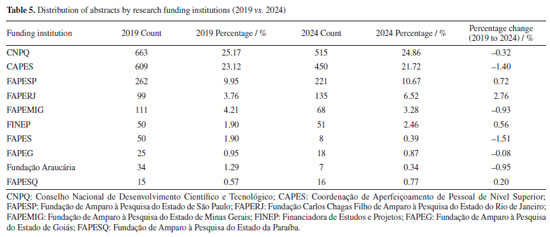
This evolution in institutional presence is closely tied to the research funding during the same period. National funding agencies such as Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), as well as the state ones, Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), Financiadora de Estudos e Projetos (FINEP), and Fundação de Amparo à Pesquisa e Inovação do Espírito Santo (FAPES), play a pivotal role in supporting research, including participation in scientific conferences.93 For instance, in Table 5 and Figures 11 and 12, FAPESP supports institutions in São Paulo, and its contribution has increased from 9.95 to 10.67%. This correlates with growth observed in São Paulo‑based universities such as USP, Unicamp (from 1.62 to 3.10%), Unesp (from 3.13 to 4.37%), and UFSCar (from 1.58 to 3.45%). The parallel increase suggests a direct influence of regional funding on institutional academic output and participation.
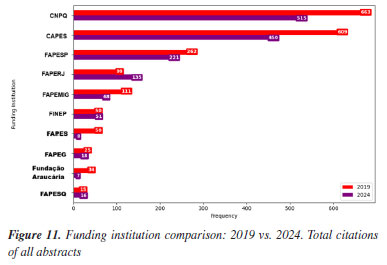

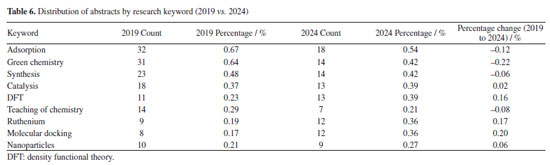
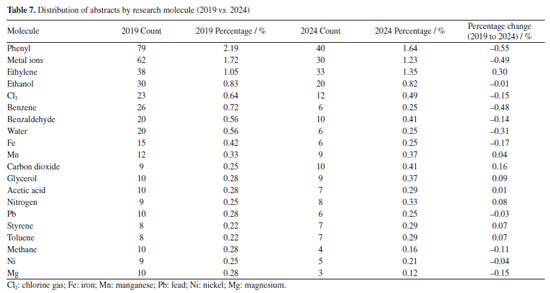
Conversely, FAPEMIG, the funding agency for Minas Gerais, experienced a decline of 0.93%, which affected the relatively stagnant growth of the UFMG participation, which increased only marginally from 2.40 to 2.47%. In an even sharper contrast, FAPES, responsible for funding in Espírito Santo, saw its share drop from 1.90 to 0.39%, coinciding with reduced visibility of institutions from that state. Other agencies showed significant positive trends. FAPERJ notably increased its share from 3.76 to 6.52%, which may have contributed to the proportional growth of Rio de Janeiro-based institutions such as UFF. This greater participation in the abstracts of the 2024 RASBQ may be associated with the significant expansion of the funding programs of FAPERJ starting in 2020. This includes initiatives such as the Cientista do Nosso Estado (CNE, Scientist from Our State), Jovem Cientista do Nosso Estado (JCNE, Young Scientist from Our State), and other scholarship programs providing associated funding.94 For instance, between 2018 and 2019, these programs awarded 379 CNE fellowships (191 in 2018 and 188 in 2019) and 271 JCNE fellowships (128 in 2018 and 143 in 2019).95 From 2020 onward, there was a marked increase in the number of fellowships, with 400 CNE and 183 JCNE awards granted in that year, reaching 446 CNE fellowships by 2023.96,97 These funding initiatives may have contributed to the maintenance and expansion of research activities within these institutions, which is reflected in the increased number of abstracts submitted by researchers from the state of Rio de Janeiro. FINEP funding also grew modestly, reflecting broader federal support for research and development. On the institutional side, smaller but steady increases were observed for UFRJ (from 3.21 to 3.33%) and UFG (from 1.75 to 1.90%). Conversely, some universities experienced declines, such as UFPR (from 2.19 to 2.01%) and, especially, UFSC, which dropped from 2.44 to 0.98%. This decline is likely related to the change in the RASBQ venue. Interestingly, this decline also mirrors the 0.55% drop in the institutional funding at UFSC, reinforcing the strong link between funding availability and research visibility. These trends highlight the dynamic interaction between financial support from public funding agencies and the academic prominence of public Brazilian universities, as well as the impact of the RASBQ venue. As funding priorities shift among federal and state agencies, so does the representation of their affiliated universities in the national scientific landscape, emphasizing the importance of sustained investment in research dissemination and in the choice of an event venue. It is also worth noting that the event held in São Paulo resulted in a smaller absolute number of abstracts from universities in the state of São Paulo compared to the event in the state of Santa Catarina. However, their percentage share of the total abstracts increased. This downward trend should be examined in other scientific events across Brazil to determine whether it reflects a broader pattern or was simply the result of an anomalous year. Keywords and molecules As shown by both keyword and molecular data from 2019 to 2024 (Tables 6 and 7; Figures 7 and 8), there is a clear and consistent shift in research priorities within the chemical sciences, marked by a rising focus on sustainability, synthesis, and computational methods. The decline of the keyword "adsorption" (from 0.67 to 0.54%) coincides with a decrease in mentions of traditional adsorption-related molecules such as "phenyl" (from 2.19 to 1.64%) and "benzene" (from 0.72 to 0.25%). These compounds, often linked to surface chemistry and organic frameworks in adsorption studies, appear to be getting less attention as the field advances toward greener and more sustainable alternatives.
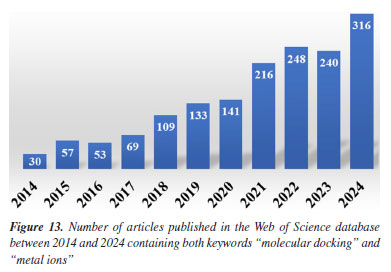
This shift is supported by the dominance of keywords such as "green chemistry" and "synthesis" in 2024, each accounting for 0.42% of mentions. This representation corresponds to growth in the relative presence of molecules such as "carbon dioxide" (0.25 to 0.41%), "glycerol" (0.28 to 0.37%), and "ethylene" (1.05 to 1.35%). These substances are frequently used in green synthetic pathways - CO2 as a reagent or reactant in carbon capture and utilization (CCU) approaches, glycerol as a bioderived solvent or platform molecule, and ethylene in sustainable polymer production - highlighting a shift toward environmentally conscious chemistry. Similarly, the notable increase in the keyword "DFT" (density functional theory, from 0.23 to 0.39%) parallels the increased usage of small, computationally tractable molecules such as glycerol, carbon dioxide, and nitrogen systems. These molecules are commonly studied in silico to model reaction mechanisms, catalyst behavior, and material properties, underscoring the growing integration of theoretical chemistry in mainstream research. The keyword "molecular docking" also doubled in presence (0.17 to 0.36%), which may help explain the increased interest in small, biologically relevant molecules such as nitrogen systems, glycerol, and acetic acid. These compounds often play key roles in docking studies, either as ligands, metabolites, or reference molecules in structure-activity relationship (SAR) analyses. The increased occurrence of the keyword "molecular docking" (from 0.17 to 0.36%) appears to be associated with the high frequency of related terms such as "metal ion", reflecting the relevance of docking approaches in biologically active systems where metal centers are often present. This trend aligns with the broader global research landscape: a search for the terms "molecular docking" and "metal ions" in the Web of Science database reveals a consistent increase in the number of publications between 2014 and 2024, indicating growing interest in the application of molecular docking techniques to metalloproteins and other bioinorganic systems (Figure 13).
Meanwhile, slight increases in the keywords "catalysis" (0.37 to 0.39%), "Mn" (0.33 to 0.37%), and "toluene" (0.22 to 0.29%) suggest a sustained, albeit evolving, interest in catalytic systems. These compounds are often involved in homogeneous or heterogeneous catalytic reactions, which remain important in both industrial applications and academic research. Taken together, these data indicate a shift away from traditional, often hazardous or less sustainable compounds, toward greener molecules and methodologies. This transformation is driven not only by environmental considerations but also by the advancement of computational tools and the broader push for innovation in chemical synthesis and molecular design. Authorship trends Figures 14 and 15 illustrate an intriguing pattern in abstract submission trends when comparing the years 2019 and 2024. Despite a considerably higher overall volume of abstracts in 2019, our analysis reveals that among the top 20, 60% of authors submitted an equal or greater number of abstracts in 2024 compared to 2019. This observation prompts further investigation into the potential factors contributing to these quantitative discrepancies.
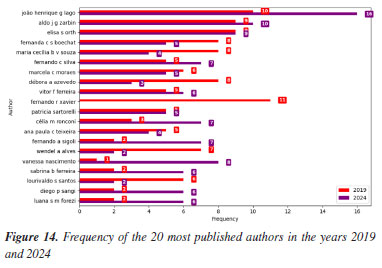

Further examination of the abstract submission data reveals that Professor João Henrique G. Lago from UFABC was the most prolific author. A significant portion of his publications feature undergraduate research (IC) and graduate (PG) students as first authors. The observed increase in submissions from 2019 to 2024 may be directly linked to the venues of RASBQ, as well as the additional funding given by FAPESP to São Paulo participants when the event takes place in the state of São Paulo. Graduate and undergraduate students often face challenges securing institutional funding for conference participation. Consequently, attending events within their state is a more viable option, even if self-funded. Undergraduate research students typically receive less institutional support compared to their graduate counterparts, as their primary focus initially lies on coursework rather than intensive research. Furthermore, the relatively lower stipends for undergraduate research scholarships compared to graduate scholarships further limit their financial independence in attending scientific events. Our analysis reveals that Aldo Jose Gorgatti Zarbin and Elisa S. Orth, both professors at the UFPR and organizers of the RASBQ 2025, are among the most prolific authors. With one additional publication in 2024, Professor Zarbin secured the second position in terms of abstract submissions, while Professor Orth holds the third position. The analysis of abstract submission trends further highlights Fernando Roberto Xavier, a professor at the UDESC. Following the publication pattern observed in the most prolific authors, Professor Xavier submitted 11 abstracts in 2019. Despite not having any publications in 2024, his total position ranks him among the most active contributors. Conversely, Vanessa do Nascimento, a professor at the IQ-UFF, demonstrates notable involvement in RASBQ in 2024, which raises a question: Did the timing of the events mainly influence her increased contributions? Or did her role as Treasurer for the SBQ during 2024-2026 play a more significant role in her decision to submit more abstracts from the work of her group to the 2024 RASBQ? Or was the funding landscape in the State of Rio in 2024 more favorable than in 2019? Or was the 2024 RASBQ venue closer to Niterói than that of 2019? This situation emphasizes the complexity of isolating single factors that influence participation choices in scientific events. These individual case studies, supported by the broader trends, collectively suggest that factors beyond mere overall abstract volume significantly impact authorship contributions. The intricate interplay of geographic accessibility, institutional support, especially for student researchers, and active involvement in scientific societies appears to be crucial in shaping submission patterns. Further comprehensive research incorporating author demographics, funding sources, and event formats would provide a more robust understanding of these complex dynamics in scientific dissemination.
CONCLUSIONS This work demonstrated how small-scale language models (SLMs) can be effective tools for extracting and organizing large volumes of text from the book of abstracts of scientific events. Despite requiring extensive post-processing, the developed model was crucial for generating structured data in CSV files containing abstract metadata, which would have been impractical to gather manually given the scale of the project. The time saved by using an SLM enabled in-depth analysis that would otherwise have required disproportionate human effort and resources. The proposed approach is based on the LLM machine learning techniques, which not only automate information extraction but also enable the application of statistical and semantic analyses that reveal significant trends. Among the most notable findings was a marked decline in the number of submitted abstracts between 2019 and 2024, likely influenced by the lingering effects of the COVID-19 pandemic, funding reductions, and contextual differences, such as the symbolic impact of the International Year of the Periodic Table in 2019. Moreover, although the 2024 event had a higher nominal budget, inflation-adjusted figures in CAPES indicate a decrease in real financial support, which affects participation rates and the dissemination of research. The comparative analysis further uncovered evolving thematic focuses: the bigram analysis, for instance, showed an increasing emphasis on synthesis coupled with characterization, as well as a growing presence of nanostructured materials and green methodologies. These patterns reflect a broader alignment between scientific innovation and environmental responsibility. Shifts in institutional participation were also observed - particularly a surprising reduction in contributions from São Paulo universities during an event held in that very state - suggesting possible regional or systemic factors that merit further investigation. Beyond these specific findings, this study highlights the broader potential of AI-powered approaches for bibliometric, historiographical, and scientific analyses. The approach outlined here is reproducible across disciplines, events, and institutions, thus offering a scalable strategy for structuring and interpreting scientific production using objective, consistent, and transparent methods. Ultimately, this work reaffirms the growing role of language models as allies in scientific analysis and production. They are not replacements for human judgment, but tools that, when critically used and statistically validated, can enhance our ability to understand and map the ever-evolving landscape of scientific knowledge.
DATA AVAILABILITY STATEMENT The source code for this work and the generated CSV file employed for all the presented analysis are available in the laboratory repository accessible at: https://github.com/Quimica-Teorica-IME.
ACKNOWLEDGMENTS I. B. thanks the Brazilian agencies Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grant number 300281/2025-0) and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ, grant number E-26/204.294/2024) for funding this research. N. M. P. R. thanks FAPERJ (grant number E-26/205.922/2022) for a post-doctoral scholarship. R. C. S. thanks Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for a PhD scholarship.
REFERENCES 1. Canal Ciência, Cesar Mansueto Giulio Lattes, https://repositorio.canalciencia.ibict.br/s/repo-cc/item/28427#?c=&m=&s=&cv=&xywh=-317%2C-23%2C1043%2C455, accessed in September 2025. 2. Mortimer, E. F.; Quim. Nova 1997, 2, 200. [Crossref] 3. Guerra, C.; Capitelli, M.; Longo, S. In The Role of Paradigms in Science: A Historical Perspective; L'Abate, L., ed.; Springer: New York, 2012. 4. Quílez, J.; Chem. Educ. Res. Pract. 2004, 5, 69. [Crossref] 5. Olawade, D. B.; Fapohunda, O.; Usman, S. O.; Akintayo, A.; Ige, A. O.; Adekunle, Y. A.; Adeola, A. O.; Chem. Afr. 2025, 8, 2707. [Crossref] 6. Baum, Z. J.; Yu, X.; Ayala, P. Y.; Zhao, Y.; Watkins, S. P.; Zhou, Q.; J. Chem. Inf. Model. 2021, 61, 3197. [Crossref] 7. Jiang, X.; Luo, S.; Liao, K.; Jiang, S.; Ma, J.; Jiang, J.; Shuai, Z.; Cell Rep. Phys. Sci. 2024, 5, 102049. [Crossref] 8. Srivastava, M.; Nandan, S.; Zaidi, A.; Samani, A. S.; Shukla, V.; Aslam, H.; Srivastava, A.; Maurya, P.; Khan, M. A.; Khan, Mohd. F.; Shanker, K.; ChemistrySelect 2025, 10, e202404446. [Crossref] 9. Rial, R. C.; Talanta 2024, 274, 125949. [Crossref] 10. Samuel, A. L.; IBM J. Res. Dev. 1959, 3, 210. [Crossref] 11. Raghavan, P.; Gayar, N.; 2019 International Conference on Computational Intelligence and Knowledge Economy (ICCIKE); Dubai, United Arab Emirates, 2019. [Crossref] 12. Gray, N. A. B.; Anal. Chim. Acta 1988, 210, 9. [Crossref] 13. Deng, W.; Breneman, C.; Embrechts, M. J.; J. Chem. Doc. 2004, 44, 699. [Crossref] 14. Wiese, B.; Omlin, C. In Innovations in Neural Information Paradigms and Applications; Bianchini, M.; Maggini, M.; Scarselli, F.; Jain, L. C., eds.; Springer: Berlin, 2009. 15. Duarte, J. C.; de Oliveira-Filho, A. G. S.; Máximo-Canadas, M.; Souza, R. C.; Borges-Junior; I.; J. Braz. Chem. Soc. 2025, 36, e-20250082. [Crossref] 16. Mousavizadegan, M.; Firoozbakhtian, A.; Hosseini, M.; Ju, H.; TrAC, Trends Anal. Chem. 2023, 167, 117216. [Crossref] 17. Prezhdo, O. V.; J. Phys. Chem. Lett. 2020, 11, 9656. [Crossref] 18. Panteleev, J.; Gao, H.; Jia, L.; Bioorg. Med. Chem. Lett. 2018, 28, 2807. [Crossref] 19. Hoerl, A. E.; Kennard, R. W.; Technometrics 1970, 12, 55. [Crossref] 20. Dempster, A. P.; Laird, N. M.; Rubin, D. B.; Journal of the Royal Statistical Society: Series B (Methodological) 1977, 39, 1. [Crossref] 21. Cover, T.; Hart, P.; IEEE Trans. Inf. Theory 1967, 13, 21. [Crossref] 22. Prati, R. C.; J. Braz. Chem. Soc. 2025, 36, e-20250067. [Crossref] 23. Le Glaz, A.; Haralambous, Y.; Kim-Dufor, D. H.; Lenca, P.; Billot, R.; Ryan, T. C.; Marsh, J.; DeVylder, J.; Walter, M.; Berrouiguet, S.; Lemey, C.; Journal of Medical Internet Research 2021, 23, e15708. [Crossref] 24. Shetty, P.; Udhayakumar, R.; Patil, A.; Manwal, M.; Vadar, P. S.; 3rd International Conference on Advancement in Electronics & Communication Engineering (AECE), Ghaziabad, India, 2023. [Crossref] 25. Bahdanau, D.; Cho, K.; Bengio, Y.; arXiv 2016. [Crossref] 26. ChatGPT, https://chat.openai.com, accessed in September 2025. 27. Goyal, P.; Pandey, S.; Jain, K.; Introduction to Natural Language Processing and Deep Learning; Apress: Berkeley, 2018. 28. Rodzin, S.; Bova, V.; Kravchenko, Y.; Rodzina, L. In Artificial Intelligence Trends in Systems; Silhavy, R., ed.; Springer: Cham, 2022, p. 121. [Crossref] 29. Liu, Y.; Zhang, M.; Computational Linguistics 2018, 44, 193. [Crossref] 30. Wu, L.; Chen, Y.; Shen, K.; Guo, X.; Gao, H.; Li, S.; Pei, J.; Long, B.; Foundations and Trends® in Machine Learning 2023, 16, 119. [Crossref] 31. Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A. N.; Kaiser, Ł.; Polosukhin, I.; arXiv 2017. [Crossref] 32. Devlin, J.; Chang, M. W.; Lee, K.; Toutanova, K.; Proceedings of the 2019 Conference of the North; Minneapolis, USA, 2019. [Crossref] 33. Gemini, https://gemini.google.com, accessed in September 2025. 34. DeepSeek-AI; Guo, D.; Yang, D.; Zhang, H.; Song, J.; Zhang, R.; Xu, R.; Zhu, Q.; Ma, S.; Wang, P.; Bi, X.; Zhang, X.; Yu, X.; Wu, Y.; Wu, Z. F.; Gou, Z.; Shao, Z.; Li, Z.; Gao, Z.; Liu, A.; Xue, B.; Wang, B.; Wu, B.; Feng, B.; Lu, C.; Zhao, C.; Deng, C.; Zhang, C.; Ruan, C.; Dai, D.; Chen, D.; Ji, D.; Li, E.; Lin, F.; Dai, F.; Luo, F.; Hao, G.; Chen, G.; Li, G.; Zhang, H.; Bao, H.; Xu, H.; Wang, H.; Ding, H.; Xin, H.; Gao, H.; Qu, H.; Li, H.; Guo, J.; Li, J.; Wang, J.; Chen, J.; Yuan, J.; Qiu, J.; Li, J.; Cai, J. L.; Ni, J.; Liang, J.; Chen, J.; Dong, K.; Hu, K.; Gao, K.; Guan, K.; Huang, K.; Yu, K.; Wang, L.; Zhang, L.; Zhao, L.; Wang, L.; Zhang, L.; Xu, L.; Xia, L.; Zhang, M.; Zhang, M.; Tang, M.; Li, M.; Wang, M.; Li, M.; Tian, N.; Huang, P.; Zhang, P.; Wang, Q.; Chen, Q.; Du, Q.; Ge, R.; Zhang, R.; Pan, R.; Wang, R.; Chen, R. J.; Jin, R. L.; Chen, R.; Lu, S.; Zhou, S.; Chen, S.; Ye, S.; Wang, S.; Yu, S.; Zhou, S.; Pan, S.; Li, S. S.; Zhou, S.; Wu, S.; Ye, S.; Yun, T.; Pei, T.; Sun, T.; Wang, T.; Zeng, W.; Zhao, W.; Liu, W.; Liang, W.; Gao, W.; Yu, W.; Zhang, W.; Xiao, W. L.; An, W.; Liu, X.; Wang, X.; Chen, X.; Nie, X.; Cheng, X.; Liu, X.; Xie, X.; Liu, X.; Yang, X.; Li, X.; Su, X.; Lin, X.; Li, X. Q.; Jin, X.; Shen, X.; Chen, X.; Sun, X.; Wang, X.; Song, X.; Zhou, X.; Wang, X.; Shan, X.; Li, Y. K.; Wang, Y. Q.; Wei, Y. X.; Zhang, Y.; Xu, Y.; Li, Y.; Zhao, Y.; Sun, Y.; Wang, Y.; Yu, Y.; Zhang, Y.; Shi, Y.; Xiong, Y.; He, Y.; Piao, Y.; Wang, Y.; Tan, Y.; Ma, Y.; Liu, Y.; Guo, Y.; Ou, Y.; Wang, Y.; Gong, Y.; Zou, Y.; He, Y.; Xiong, Y.; Luo, Y.; You, Y.; Liu, Y.; Zhou, Y.; Zhu, Y. X.; Xu, Y.; Huang, Y.; Li, Y.; Zheng, Y.; Zhu, Y.; Ma, Y.; Tang, Y.; Zha, Y.; Yan, Y.; Ren, Z. Z.; Ren, Z.; Sha, Z.; Fu, Z.; Xu, Z.; Xie, Z.; Zhang, Z.; Hao, Z.; Ma, Z.; Yan, Z.; Wu, Z.; Gu, Z.; Zhu, Z.; Liu, Z.; Li, Z.; Xie, Z.; Song, Z.; Pan, Z.; Huang, Z.; Xu, Z.; Zhang, Z.; Zhang, Z.; arXiv 2025. [Crossref] 35. Brown, T. B.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; Agarwal, S.; Herbert-Voss, A.; Krueger, G.; Henighan, T.; Child, R.; Ramesh, A.; Ziegler, D. M.; Wu, J.; Winter, C.; Hesse, C.; Chen, M.; Sigler, E.; Litwin, M.; Gray, S.; Chess, B.; Clark, J.; Berner, C.; Mccandlish, S.; Radford, A.; Sutskever, I.; Amodei, D.; arXiv 2020. [Crossref] 36. Raza, M.; Jahangir, Z.; Riaz, M. B.; Saeed, M. J.; Sattar, M. A.; Sci. Rep. 2025, 15, 13755. [Crossref] 37. Mohan, G. B.; Kumar, R. P.; Krishh, P. V.; Keerthinathan, A.; Lavanya, G.; Meghana, M. K. U.; Sulthana, S.; Doss, S.; Knowledge and Information Systems 2024, 66, 5047. [Crossref] 38. Yang, R.; Tan, T. F.; Lu, W.; Thirunavukarasu, A. J.; Ting, D. S. W.; Liu, N.; Health Care Science 2023, 2, 255. [Crossref] 39. Bommasani, R.; Hudson, D. A.; Adeli, E.; Altman, R.; Arora, S.; von Arx, S.; Bernstein, M. S.; Bohg, J.; Bosselut, A.; Brunskill, E.; Brynjolfsson, E.; Buch, S.; Card, D.; Castellon, R.; Chatterji, N.; Chen, A.; Creel, K.; Davis, J. Q.; Demszky, D.; Donahue, C.; Doumbouya, M.; Durmus, E.; Ermon, S.; Etchemendy, J.; Ethayarajh, K.; Fei-Fei, L.; Finn, C.; Gale, T.; Gillespie, L.; Goel, K.; Goodman, N.; Grossman, S.; Guha, N.; Hashimoto, T.; Henderson, P.; Hewitt, J.; Ho, D. E.; Hong, J.; Hsu, K.; Huang, J.; Icard, T.; Jain, S.; Jurafsky, D.; Kalluri, P.; Karamcheti, S.; Keeling, G.; Khani, F.; Khattab, O.; Koh, P. W.; Krass, M.; Krishna, R.; Kuditipudi, R.; Kumar, A.; Ladhak, F.; Lee, M.; Lee, T.; Leskovec, J.; Levent, I.; Li, X. L.; Li, X.; Ma, T.; Malik, A.; Manning, C. D.; Mirchandani, S.; Mitchell, E.; Munyikwa, Z.; Nair, S.; Narayan, A.; Narayanan, D.; Newman, B.; Nie, A.; Niebles, J. C.; Nilforoshan, H.; Nyarko, J.; Ogut, G.; Orr, L.; Papadimitriou, I.; Park, J. S.; Piech, C.; Portelance, E.; Potts, C.; Raghunathan, A.; Reich, R.; Ren, H.; Rong, F.; Roohani, Y.; Ruiz, C.; Ryan, J.; Ré, C.; Sadigh, D.; Sagawa, S.; Santhanam, K.; Shih, A.; Srinivasan, K.; Tamkin, A.; Taori, R.; Thomas, A. W.; Tramèr, F.; Wang, R. E.; Wang, W.; Wu, B.; Wu, J.; Wu, Y.; Xie, S. M.; Yasunaga, M.; You, J.; Zaharia, M.; Zhang, M.; Zhang, T.; Zhang, X.; Zhang, Y.; Zheng, L.; Zhou, K.; Liang, P.; arXiv 2022. [Crossref] 40. Meyer, J. G.; Urbanowicz, R. J.; Martin, P. C. N.; O'Connor, K.; Li, R.; Peng, P. C.; Bright, T. J.; Tatonetti, N.; Won, K. J.; Gonzalez-Hernandez, G.; Moore, J. H.; BioData Min. 2023, 16, 20. [Crossref] 41. Mishra, T.; Sutanto, E.; Rossanti, R.; Pant, N.; Ashraf, A.; Raut, A.; Uwabareze, G.; Oluwatomiwa, A.; Zeeshan, B.; Sci. Rep. 2024, 14, 31672. [Crossref] 42. Jiang, X.; Dong, Y.; Wang, L.; Fang, Z.; Shang, Q.; Li, G.; Jin, Z.; Jiao, W.; ACM Transactions on Software Engineering and Methodology 2024, 33, 1. [Crossref] 43. Xu, F.; Hao, Q.; Zong, Z.; Wang, J.; Zhang, Y.; Wang, J.; Lan, X.; Gong, J.; Ouyang, T.; Meng, F.; Shao, C.; Yan, Y.; Yang, Q.; Song, Y.; Ren, S.; Hu, X.; Li, Y.; Feng, J.; Gao, C.; Li, Y.; arXiv 2025. [Crossref] 44. Martins, M. R.; World J. Adv. Res. Rev. 2023, 20, 1898. [Crossref] 45. Magister, L. C.; Mallinson, J.; Adamek, J.; Malmi, E.; Severyn, A.; arXiv 2023. [Crossref] 46. Ollama, https://ollama.com, accessed in September 2025. 47. Anand, Y.; Nussbaum, Z.; Duderstadt, B.; Schmidt, B.; Mulyar, A.; GPT4All: Run Local LLMs on Any Device, https://github.com/nomic-ai/gpt4all, accessed in September 2025. 48. Touvron, H.; Lavril, T.; Izacard, G.; Martinet, X.; Lachaux, M. A.; Lacroix, T.; Rozière, B.; Goyal, N.; Hambro, E.; Azhar, F.; Rodriguez, A.; Joulin, A.; Grave, E.; Lample, G.; arXiv 2023. [Crossref] 49. Dettmers, T.; Pagnoni, A.; Holtzman, A.; Zettlemoyer, L.; arXiv 2023. [Crossref] 50. Miah, M. S. U.; Kabir, M. M.; Sarwar, T. B.; Safran, M.; Alfarhood, S.; Mridha, M. F.; Sci. Rep. 2024, 14, 9603. [Crossref] 51. Yin, Y. J.; Chen, B. Y.; Chen, B.; arXiv 2024. [Crossref] 52. Zhang, J.; Zhao, Y.; Saleh, M.; Liu, P. J.; arXiv 2020. [Crossref] 53. Belcak, P.; Heinrich, G.; Diao, S.; Fu, Y.; Dong, X.; Muralidharan, S.; Lin, Y. C.; Molchanov, P.; arXiv 2025. [Crossref] 54. González, C. J.; Bugarín-Diz, A.; Alonso-Moral, J.; Taboada, J.; Proceedings of the 15th International Conference on Natural Language Generation; Waterville, USA, 2022. [Crossref] 55. Tonmoy, S. M. T. I.; Zaman, S. M. M.; Jain, V.; Rani, A.; Rawte, V.; Chadha, A.; Das, A.; arXiv 2024. [Crossref] 56. Sachdeva, N.; Coleman, B.; Kang, W. C.; Ni, J.; Hong, L.; Chi, E. H.; Caverlee, J.; McAuley, J.; Cheng, D. Z.; arXiv 2024. [Crossref] 57. Stuyck, T.; Van Lith, P.; Demeester, E.; SSRN 2024. [Crossref] 58. Jafari, M.; Tao, X.; Barua, P.; Tan, R. S.; Acharya, U. R.; Information Fusion 2025, 118, 102982. [Crossref] 59. Zhang, Y.; Ye, Z.; Hu, D.; Qi, S.; Sun, Z.; Yang, J.; Ma, Y.; Zhang, Y.; Zhang, J.; Li, Z.; ChemRxiv 2025. [Crossref] 60. Bechara, E. J. H.; Viertler, H.; Quim. Nova 1997, 20, 63. [Crossref] 61. Química Nova, https://quimicanova.sbq.org.br, accessed in September 2025. 62. Sociedade Brasileira de Química (SBQ), Reuniões Anuais da SBQ, https://www.sbq.org.br/reunioes-anuais, accessed in September 2025. 63. Sociedade Brasileira de Química (SBQ), Reuniões Anuais da SBQ, RASBQ (2023-2025): Resumos e Programa, https://www.sbq.org.br/rasbq-resumos-e-programas-2, accessed in September 2025. 64. Schimmenti, A.; Pasqual, V.; Tomasi, F.; Vitali, F.; van Erp, M.; arXiv 2024. [Crossref] 65. Li, L.; arXiv 2024. [Crossref] 66. Litaina, T.; Soularidis, A.; Bouchouras, G.; Kotis, K.; Kavakli, E.; First International Workshop of Semantic Digital Humanities; Hersonissos, Greece, 2024. [Link] accessed in September 2025 67. Ji, X.; Wu, X.; Deng, R.; Yang, Y.; Wang, A.; Zhu, Y.; J. Environ. Manage. 2025, 373, 123667. [Crossref] 68. PyMuPDF, https://pymupdf.readthedocs.io/en/latest/the-basics.html#supported-file-types, accessed in September 2025. 69. Borges, P.; Ladele, A.; Cunha, Y.; Moraes, D.; Costa, P.; Santos, P.; Rocha, R.; Busson, A.; Duarte, J.; Colcher, S.; XX Simpósio Brasileiro de Sistemas Multimídia e Web: Minicursos; Juiz de Fora, Brasil, 2024. [Crossref] 70. Gemma Team; Riviere, M.; Pathak, S.; Sessa, P. G.; Hardin, C.; Bhupatiraju, S.; Hussenot, L.; Mesnard, T.; Shahriari, B.; Ramé, A.; Ferret, J.; Liu, P.; Tafti, P.; Friesen, A.; Casbon, M.; Ramos, S.; Kumar, R.; Le Lan, C.; Jerome, S.; Tsitsulin, A.; Vieillard, N.; Stanczyk, P.; Girgin, S.; Momchev, N.; Hoffman, M.; Thakoor, S.; Grill, J. B.; Neyshabur, B.; Bachem, O.; Walton, A.; Severyn, A.; Parrish, A.; Ahmad, A.; Hutchison, A.; Abdagic, A.; Carl, A.; Shen, A.; Brock, A.; Coenen, A.; Laforge, A.; Paterson, A.; Bastian, B.; Piot, B.; Wu, B.; Royal, B.; Chen, C.; Kumar, C.; Perry, C.; Welty, C.; Choquette-Choo, C. A.; Sinopalnikov, D.; Weinberger, D.; Vijaykumar, D.; Rogozińska, D.; Herbison, D.; Bandy, E.; Wang, E.; Noland, E.; Moreira, E.; Senter, E.; Eltyshev, E.; Visin, F.; Rasskin, G.; Wei, G.; Cameron, G.; Martins, G.; Hashemi, H.; Klimczak-Plucińska, H.; Batra, H.; Dhand, H.; Nardini, I.; Mein, J.; Zhou, J.; Svensson, J.; Stanway, J.; Chan, J.; Zhou, J. P.; Carrasqueira, J.; Iljazi, J.; Becker, J.; Fernandez, J.; van Amersfoort, J.; Gordon, J.; Lipschultz, J.; Newlan, J.; Ji, J.; Mohamed, K.; Badola, K.; Black, K.; Millican, K.; McDonell, K.; Nguyen, K.; Sodhia, K.; Greene, K.; Sjoesund, L. L.; Usui, L.; Sifre, L.; Heuermann, L.; Lago, L.; McNealus, L.; Soares, L. B.; Kilpatrick, L.; Dixon, L.; Martins, L.; Reid, M.; Singh, M.; Iverson, M.; Görner, M.; Velloso, M.; Wirth, M.; Davidow, M.; Miller, M.; Rahtz, M.; Watson, M.; Risdal, M.; Kazemi, M.; Moynihan, M.; Zhang, M.; Kahng, M.; Park, M.; Rahman, M.; Khatwani, M.; Dao, N.; Bardoliwalla, N.; Devanathan, N.; Dumai, N.; Chauhan, N.; Wahltinez, O.; Botarda, P.; Barnes, P.; Barham, P.; Michel, P.; Jin, P.; Georgiev, P.; Culliton, P.; Kuppala, P.; Comanescu, R.; Merhej, R.; Jana, R.; Rokni, R. A.; Agarwal, R.; Mullins, R.; Saadat, S.; Carthy, S. M.; Cogan, S.; Perrin, S.; Arnold, S. M. R.; Krause, S.; Dai, S.; Garg, S.; Sheth, S.; Ronstrom, S.; Chan, S.; Jordan, T.; Yu, T.; Eccles, T.; Hennigan, T.; Kocisky, T.; Doshi, T.; Jain, V.; Yadav, V.; Meshram, V.; Dharmadhikari, V.; Barkley, W.; Wei, W.; Ye, W.; Han, W.; Kwon, W.; Xu, X.; Shen, Z.; Gong, Z.; Wei, Z.; Cotruta, V.; Kirk, P.; Rao, A.; Giang, M.; Peran, L.; Warkentin, T.; Collins, E.; Barral, J.; Ghahramani, Z.; Hadsell, R.; Sculley, D.; Banks, J.; Dragan, A.; Petrov, S.; Vinyals, O.; Dean, J.; Hassabis, D.; Kavukcuoglu, K.; Farabet, C.; Buchatskaya, E.; Borgeaud, S.; Fiedel, N.; Joulin, A.; Kenealy, K.; Dadashi, R.; Andreev, A.; arXiv 2024. [Crossref] 71. Llama3.1, https://ollama.com/library/llama3.1, accessed in September 2025. 72. Llama3.2, Model Card and Prompt Formats, https://www.llama.com/docs/model-cards-and-prompt-formats/llama3_2/, accessed in September 2025. 73. Abdin, M.; Aneja, J.; Awadalla, H.; Awadallah, A.; Awan, A. A.; Bach, N.; Bahree, A.; Bakhtiari, A.; Bao, J.; Behl, H.; Benhaim, A.; Bilenko, M.; Bjorck, J.; Bubeck, S.; Cai, M.; Cai, Q.; Chaudhary, V.; Chen, D.; Chen, D.; Chen, W.; Chen, Y. C.; Chen, Y. L.; Cheng, H.; Chopra, P.; Dai, X.; Dixon, M.; Eldan, R.; Fragoso, V.; Gao, J.; Gao, M.; Gao, M.; Garg, A.; Del Giorno, A.; Goswami, A.; Gunasekar, S.; Haider, E.; Hao, J.; Hewett, R. J.; Hu, W.; Huynh, J.; Iter, D.; Jacobs, S. A.; Javaheripi, M.; Jin, X.; Karampatziakis, N.; Kauffmann, P.; Khademi, M.; Kim, D.; Kim, Y. J.; Kurilenko, L.; Lee, J. R.; Lee, Y. T.; Li, Y.; Li, Y.; Liang, C.; Liden, L.; Lin, X.; Lin, Z.; Liu, C.; Liu, L.; Liu, M.; Liu, W.; Liu, X.; Luo, C.; Madan, P.; Mahmoudzadeh, A.; Majercak, D.; Mazzola, M.; Mendes, C. C. T.; Mitra, A.; Modi, H.; Nguyen, A.; Norick, B.; Patra, B.; Perez-Becker, D.; Portet, T.; Pryzant, R.; Qin, H.; Radmilac, M.; Ren, L.; de Rosa, G.; Rosset, C.; Roy, S.; Ruwase, O.; Saarikivi, O.; Saied, A.; Salim, A.; Santacroce, M.; Shah, S.; Shang, N.; Sharma, H.; Shen, Y.; Shukla, S.; Song, X.; Tanaka, M.; Tupini, A.; Vaddamanu, P.; Wang, C.; Wang, G.; Wang, L.; Wang, S.; Wang, X.; Wang, Y.; Ward, R.; Wen, W.; Witte, P.; Wu, H.; Wu, X.; Wyatt, M.; Xiao, B.; Xu, C.; Xu, J.; Xu, W.; Xue, J.; Yadav, S.; Yang, F.; Yang, J.; Yang, Y.; Yang, Z.; Yu, D.; Yuan, L.; Zhang, C.; Zhang, C.; Zhang, J.; Zhang, L. L.; Zhang, Y.; Zhang, Y.; Zhang, Y.; Zhou, X.; arXiv 2024. [Crossref] 74. Abdin, M.; Agarwal, S.; Awadallah, A.; Balachandran, V.; Behl, H.; Chen, L.; De Rosa, G.; Gunasekar, S.; Javaheripi, M.; Joshi, N.; Kauffmann, P.; Lara, Y.; César, C.; Mendes, T.; Mitra, A.; Nushi, B.; Papailiopoulos, D.; Saarikivi, O.; Shah, S.; Shrivastava, V.; Vineet, V.; Wu, Y.; Yousefi, S.; Zheng, G.; arXiv 2025. [Crossref] 75. Souza Júnior, R. C.; Borges Júnior, I.; https://github.com/Quimica-Teorica-IME/SBQ-SLM-Trends-Analysis, accessed in September 2025. 76. Sociedade Brasileira de Química, Anais da 43a Reunião Anual da Sociedade Brasileira de Química (RASBQ 2020), https://proceedings.science/rasbq-2020/trabalhos?lang=pt-br, accessed in September 2025. 77. Sociedade Brasileira de Química, 44a Reunião Anual da Sociedade Brasileira de Química (RASBQ 2021), https://eventos.galoa.com.br/rasbq-2021/page/725-home, accessed in September 2025. 78. Agência gov, Execução Orçamentária da Capes é a Maior dos Últimos 7 Anos, https://agenciagov.ebc.com.br/noticias/202404/execucao-orcamentaria-da-capes-e-a-maior-dos-ultimos-7-anos, accessed in September 2025. 79. Gov.br, CAPES concedeu quase 200 mil Bolsas em 2024, https://www.gov.br/capes/pt-br/assuntos/noticias/capes-concedeu-quase-200-mil-bolsas-em-2024, accessed in September 2025. 80. United Nations Educational, Scientific and Cultural Organization (UNESCO), The International Year of the Periodic Table of Chemical Elements, Global Report 2019, https://unesdoc.unesco.org/ark:/48223/pf0000374793.locale=en, accessed in September 2025. 81. Sun, H.; Liu, S.; Zhou, G.; Ang, H. M.; Tadé, M. O.; Wang, S.; ACS Appl. Mater. Interfaces 2012, 4, 5466. [Crossref] 82. Mitsudome, T.; Kaneda, K.; Green Chem. 2013, 15, 2636. [Crossref] 83. Anjali; Mishra, A.; Khurana, M.; Pani, B.; Awasthi, S. K.; ChemistrySelect 2025, 10, e202404742. [Crossref] 84. Badoni, A.; Thakur, S.; Vijayan, N.; Swart, H. C.; Bechelany, M.; Chen, Z.; Sun, S.; Cai, Q.; Chen, Y.; Prakash, J.; Catal. Sci. Technol. 2025, 15, 1702. [Crossref] 85. Grisel, R.; Weststrate, K. J.; Gluhoi, A.; Nieuwenhuys, B. E.; Gold Bull. 2002, 35, 39. [Crossref] 86. Kirubakaran, D.; Wahid, J. B. A.; Karmegam, N.; Jeevika, R.; Sellapillai, L.; Rajkumar, M.; SenthilKumar, K. J.; Biomed. Mater. Devices 2026, 4, 338. [Crossref] 87. Aggarwal, R.; Sheikh, A.; Akhtar, M.; Ghazwani, M.; Hani, U.; Sahebkar, A.; Kesharwani, P.; Mol. Cancer 2025, 24, 88. [Crossref] 88. Sun, M.; Wang, S.; Liang, Y.; Wang, C.; Zhang, Y.; Liu, H.; Zhang, Y.; Han, L.; Nano-Micro Lett. 2025, 17, 34. [Crossref] 89. Seth, A.; Mandal, P.; Hitaishi, P.; Giri, R. P.; Murphy, B. M.; Ghosh, S. K.; Phys. Chem. Chem. Phys. 2025, 27, 1884. [Crossref] 90. Sundar, L. S.; Ashraf, M. W.; Inorg. Chem. Commun. 2025, 174, 113936. [Crossref] 91. Khan, M. B.; Parvaz, M.; Khan, Z. H.; Recent Trends in Nanomaterials: Synthesis and Properties; Springer: Singapore, 2017. 92. Verma, H. N.; Singh, P.; Chavan, R. M.; Vet. World 2014, 7, 72. [Crossref] 93. Scartassini, V. B.; de Moura, A. M. M.; Revista Ibero-Americana de Ciência da Informação 2020, 13, 915. [Crossref] 94. Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Programas, https://www.faperj.br/?id=31.5.4, accessed in September 2025. 95. Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), FAPERJ Divulga o Resultado dos Programas Cientista e Jovem Cientista do Nosso Estado, https://siteantigo.faperj.br/?id=3628.2.4, accessed in September 2025. 96. Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Anunciado Resultado Preliminar da Edição 2023 dos Programas Cientista e Jovem Cientista do Nosso Estado, https://www.faperj.br/?id=602.7.0, accessed in September 2025. 97. Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), FAPERJ Divulga Resultado dos Editais CNE e JCNE, https://siteantigo.faperj.br/?id=3836.2.4, accessed in September 2025. |
On-line version ISSN 1678-7064 Printed version ISSN 0100-4042
Qu�mica Nova
Publica��es da Sociedade Brasileira de Qu�mica
Caixa Postal: 26037
05513-970 S�o Paulo - SP
Tel/Fax: +55.11.3032.2299/+55.11.3814.3602
Free access